STAGE
Spatiotemporal Gene Expression Atlas of Autism
Autism is a highly heritable neurodevelopmental condition that is understood as a diverse spectrum of characteristics. In over 10% of autistic people, rare and de novo loss-of-functions mutations strongly predispose to profound autism and co-occurring developmental disorders and intellectual disabilities. Understanding how variants converge on specific brain circuits and cell populations during human brain development, particularly in less explored brain areas, will be crucial to understand the etiology of profound autism.
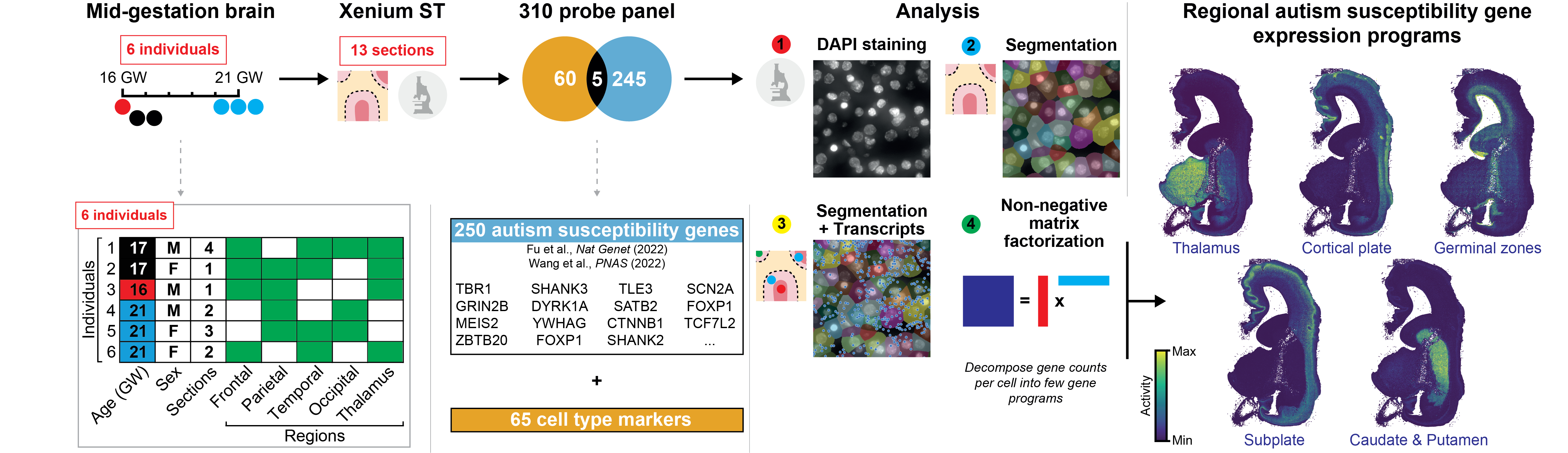
Supported by the Simons Foundation Autism Research Initiative, we charted the spatiotemporal expression patterns of 250 such autism susceptibility genes with characterized mutations, across the second trimester developing human forebrain. Profiling over 10 million cells, we found convergence of these genes across a small number of brain regional programs. The resulting STAGE atlas uniquely combines:
- Single-cell resolution spatial gene expression of >10 million cells across diverse human brain regions at 3 timepoints in the second trimester.
- Expert-annotated neuroanatomy and cell states to contextualize where genes are expressed.
- State-of-the-art computational analyses to uncover co-expressing gene programs.
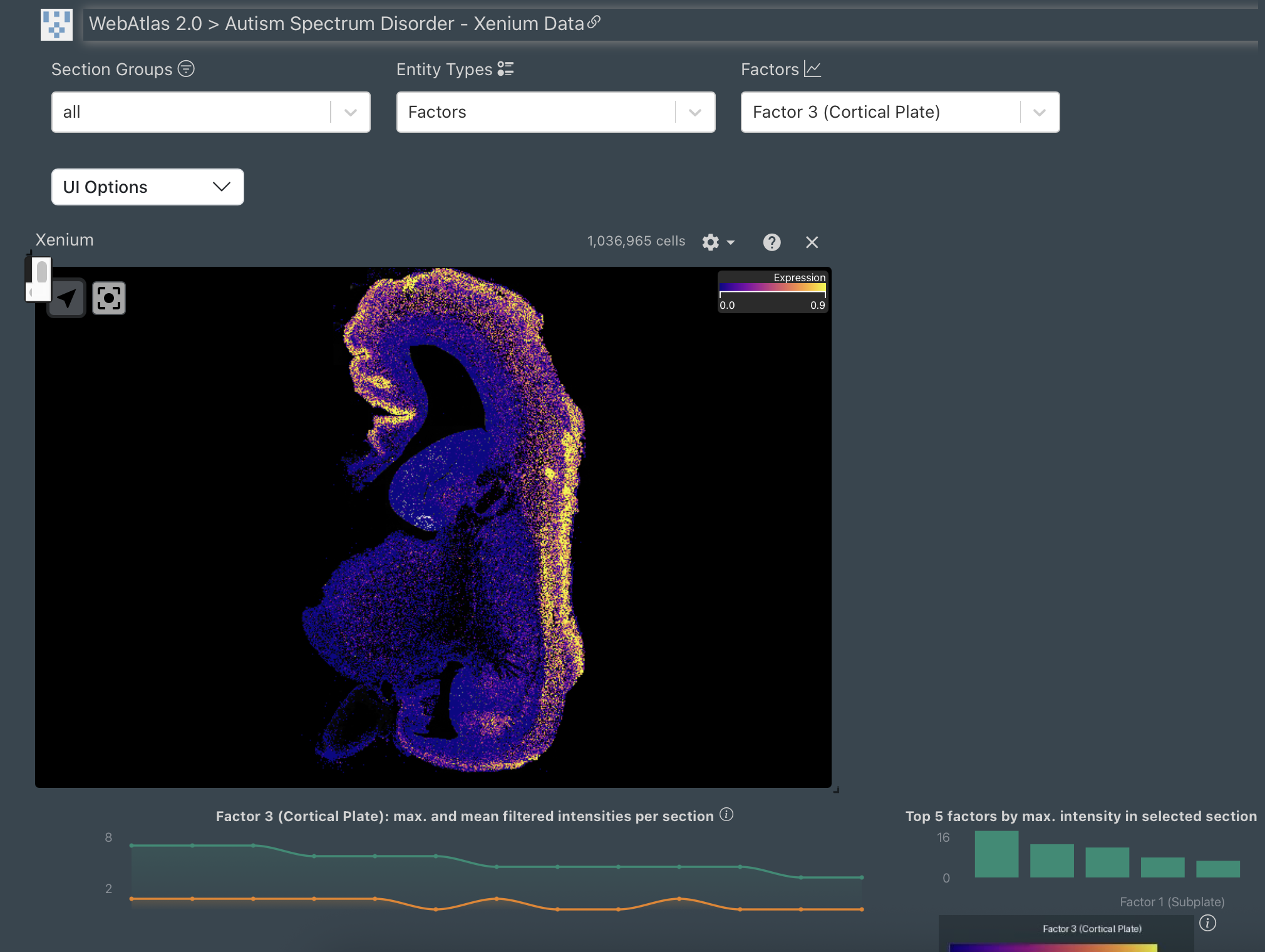
The initial spatial transcriptomics dataset can be interactively navigated and downloaded on this portal.
Publications
A spatial transcriptomic atlas of autism-associated genes identifies convergence in the developing human thalamus
Aivazidis A, Memi F, Rademaker K, et al.
Preprint available https://www.biorxiv.org/content/10.1101/2025.11.05.685843v1Datasets
Spatial data on Webatlas
10.8 million single cell and spatially resolved transcriptomes. The resulting data shows gene expression, brain regional annotation, and mapped cell types for each spatial tissue location.
Our Team

Omer
Bayraktar
Principal Investigator
Wellcome Sanger Institute

Alexander
Aivazidis
Omics Computational Analysis
Wellcome Sanger Institute

Fani
Memi
Spatial Transcriptomics Specialist
Wellcome Sanger Institute

Koen
Rademaker
Omics Computational Analysis
Wellcome Sanger Institute

Mahmoud
Koko
Genetic Analysis
Wellcome Sanger Institute

Kenny
Roberts
Spatial Transcriptomics Data Generation
Wellcome Sanger Institute

Andrew
Trinh
Spatial Transcriptomics Data Generation
Wellcome Sanger Institute

Robert
Petryszak
Data Visualization Portal
Wellcome Sanger Institute

Vitalii
Kleshchevnikov
Omics Computational Analysis
Wellcome Sanger Institute

Elizabeth
Tuck
Histology specialist
Wellcome Sanger Institute

Steven
Lisgo
Tissue Sampling
Human Developmental Biology Resource

Tong
Li
Imaging Analysis
Wellcome Sanger Institute

Stanislaw
Makarchuk
Imaging Analysis
Wellcome Sanger Institute

Tomasz
Nowakowski
Co-Principal Investigator
University of California, San Francisco

Hilary
Martin
Co-Principal Investigator
Wellcome Sanger Institute
Acknowledgements
Wellcome Sanger Institute; Human Developmental Biology Resource; Newcastle University; University of California, San Francisco; Simons Foundation Autism Research Initiative (SFARI). Banner image from K. Roberts and A. Trinh.

